Lipidomics
See Figure 2 for the meanings of the abbreviations used in this chapter.
1. DI-MS/MS (DDA)
A project dealing with data dependent DI-MS acquisition in combination with the in silico retention time and MS/MS databases for lipids is demonstrated.
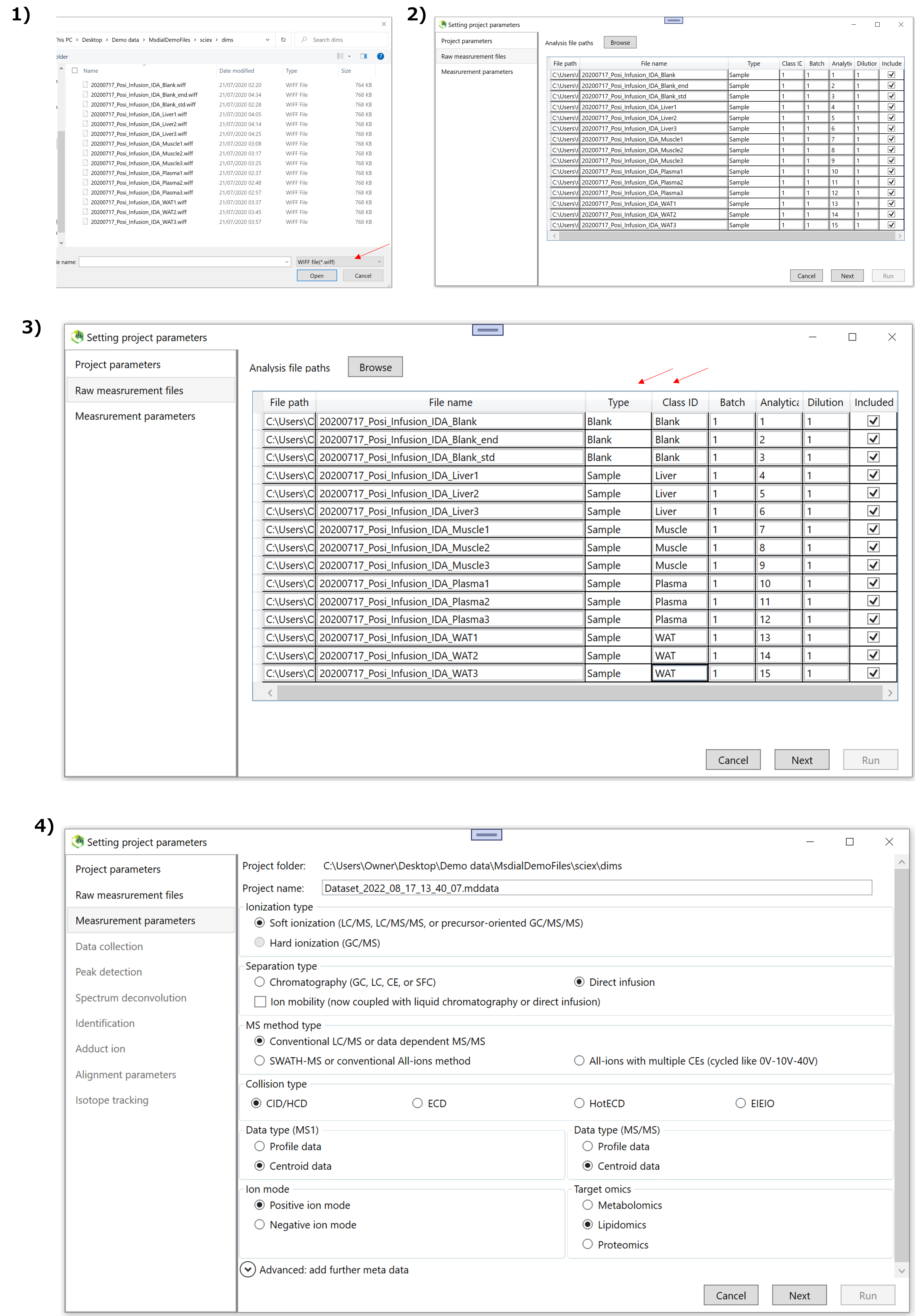
This tutorial uses 15 demonstration files which are downloadable from the below link.
http://prime.psc.riken.jp/compms/msdial/download/demo/dims.zip
Experiment summary:
The direct infusion MS data analyzing plasma, white adipose tissue (WAT), muscle, and liver of mice in addition to the procedure blank samples. The MS/MS spectrum was acquired by information dependent acquisition (IDA) mode of SCIEX meaning data dependent acquisition (DDA).
1-1 Starting up your project

To see the above operation in YouTube, check out the following
2. DI-MS/MS (MSall)
A project dealing with MSall DI-MS acquisition in combination with the in silico retention time and MS/MS databases for lipids is demonstrated.
Additionally, you can watch our tutorial video:
3. DI-MS/MS (DDA)
A project dealing with DDA DI-MS acquisition in combination with the in silico retention time and MS/MS databases for lipids is demonstrated.
Additionally, you can watch our tutorial video:
4. LC-MS/MS (SWATH)
A project dealing with data independent LC-MS acquisition using the Sequential Window Acquisition of all Theoretical Mass Spectra (SWATH-MS) in combination with the in silico retention time and MS/MS databases for lipids is demonstrated.
Input Data
This tutorial uses 23 demonstration files which are downloadable from http://prime.psc.riken.jp/archives/data/DropMet/019/20140809_MSDIAL_DemoFiles_Swath(abf).zip. The experimental protocol is described in previous research: http://www.nature.com/nmeth/journal/v12/n6/abs/nmeth.3393.html.
Experiment summary:
Liquid chromatography: total 15 min run per sample with Waters Acquity UPLC CSH C18 column (100×2.1 mm; 1.7 μm). Mass spectrometer: SWATH method with negative ion mode.
- MS1 accumulation time, 100 ms
- MS2 accumulation time, 10 ms
- Collision energy, 45 V
- Collision energy spread, 15 V
- Cycle time, 731 ms
- Q1 window, 21 Da
- Mass range, m/z 100-1250
- Modifier, Ammonium formate (HCOONH4)
Additionally, you can watch our tutorial video:
5. LC-MS/MS (Hybrid MS-DDA/SWATH)
A project dealing with LC-MS/MS (Hybrid MS-DDA/SWATH) acquisition in combination with the in silico retention time and MS/MS databases for lipids is demonstrated.
Hybrid MS-DDA/SWATH is a novel strategy combining DIA and DDA in fast LC-MS-based analytical batches, intending to increase annotation rates.
The input data to the workflow is located in .
See the Zenodo dataset for description of the input data.
The Workflow
- Download and unzip the tutorial dataset.
- Add the LBM2 file to the MS-DIAL folder.
NOTE: Exclude the original LBM2 format. The program accepts only one LBM/LBM2 file in the MS-DIAL folder. - Start up a new project in MS-DIAL.
- Follow the YouTube video below to operate the MS-DIAL5 GUI.
- The output is obtained as *.mdproject.
Reference
See below for the details of “Hybrid MS”.
K. Tokiyoshi et al. Anal Chem, 96, 991-996. 2024
6. LC-MS/MS (EAD) - deep structural lipid annotation
The input data to the workflow is located in .
See the Zenodo dataset for description of the input data.
Additionally, you can watch our tutorial video:
7. DI-IM-MS/MS
A project dealing with DI-IM-MS acquisition in combination with the in silico retention time and MS/MS databases for lipids is demonstrated.
Additionally, you can watch our tutorial video:
8. LC-IM-MS/MS(SWATH)
A project dealing with LC-IM-MS/MS acquisition in combination with the in silico retention time and MS/MS databases for lipids is demonstrated.
Additionally, you can watch our tutorial video: